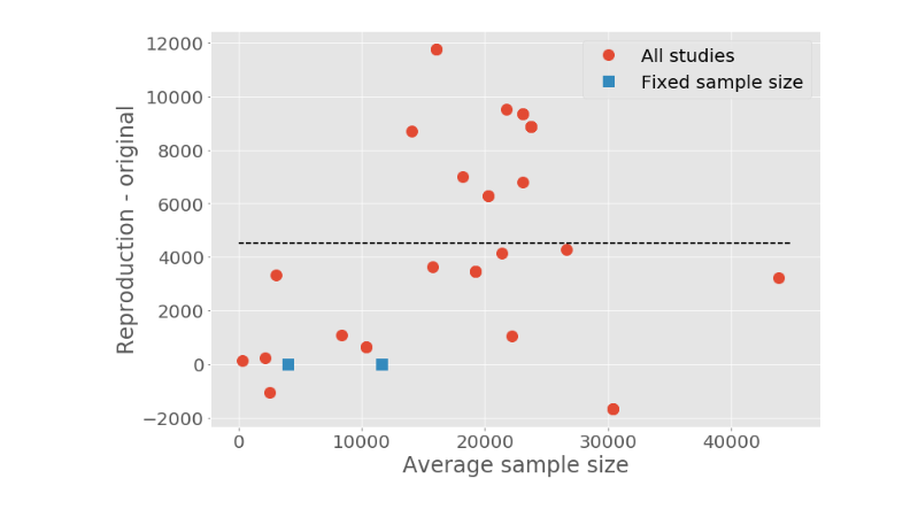
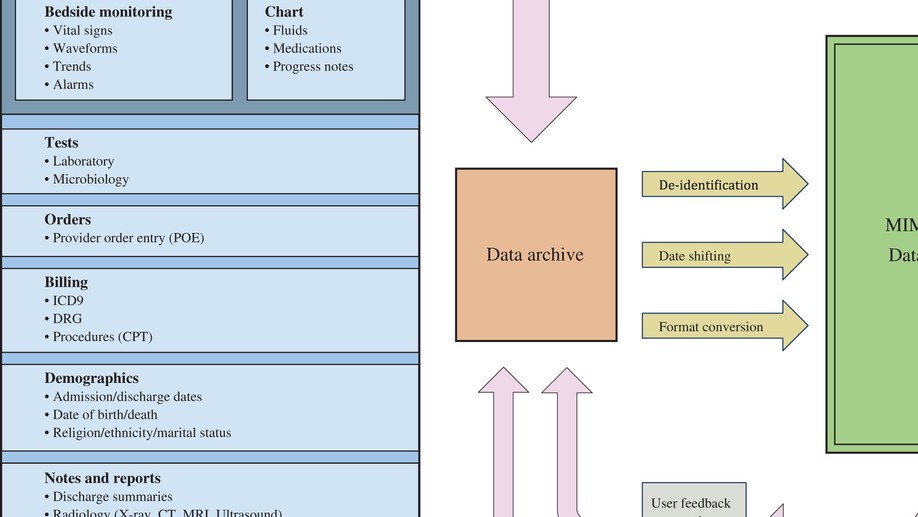
The number of researcher degrees of freedom that contribute to the performance of a model presents a challenge when seeking to compare reported performance of such models. We review publications that have reported performance of mortality prediction models based on the Medical Information Mart for Intensive Care (MIMIC) database and attempted to reproduce datasets for 38 experiments corresponding to 28 published studies. In half of the experiments, the sample size we acquired was 25% greater or smaller than the sample size reported. While accurate reproduction of each study cannot be guaranteed, we believe that these results highlight the need for more consistent reporting of model design and methodology to allow performance improvements to be compared. We discuss the challenges in reproducing the cohorts used in the studies, highlighting the importance of clearly reported methods (e.g. data cleansing, variable selection, cohort selection) and the need for open code and publicly available benchmarks.